A Glimpse in the Dark: Proteins Under the Microscope

An important element of research in drug design is being able to see the structural changes in a particle when it’s bound to something versus when it’s not. But using electron microscopy on such particles—one standard approach to study their structure—presents a challenge: the electron beams immediately destroy the structure of whatever you’re trying to look at.
“You only get a glimpse,” explains Alp Kucukelbir, a doctoral candidate in Biomedical Engineering. “It’s like entering a room and looking around for just a second before the lights are turned out, and being asked to reconstruct what the room looked like.”
Many algorithms exist to perform single particle reconstruction, the process by which researchers build a 3D image of the particle after its structure has been destroyed. But current methods create very noisy images, and require additional manual filtering to separate the particle from the background in the resulting reconstruction.
Kucukelbir, working with Fred Sigworth, Professor of Physiology & Biomedical Engineering, and Hemant Tagare, Associate Professor of Diagnostic Radiology, Electrical Engineering & Biomedical Engineering, has developed a new algorithm for single particle reconstruction, which uses an adaptive process to determine what the particle looks like and differentiate it from the noise in the images.
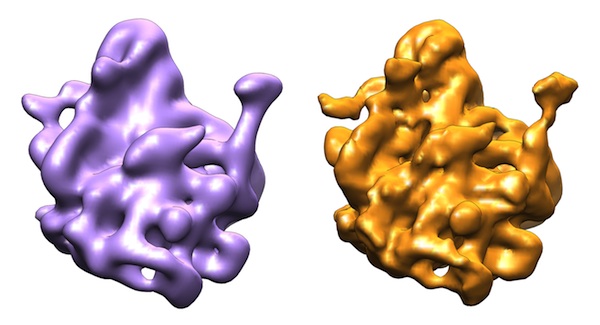 |
| Left: A reconstruction using traditional methods (in purple). Right: A reconstruction using the Yale researchers' method (orange), which offers increased resolution, and therefore more detail. |
“This approach automatically suppresses the ‘junk’ that comes from noise in the reconstruction,” says Kucukelbir. In addition, he explains, in contrast to other methods that require parameters to be manually set, “All of the algorithm’s parameters are determined automatically from actual data.” The resulting reconstruction has an improved resolution and signal-to-noise ratio compared to those created using existing algorithms, and automatically separates the particle from the background with no additional filtering required.
In their future work, the group hopes to incorporate additional information into their algorithm for even better resolution and suppression of noise.
“When we’re doing a reconstruction, we know that we’re reconstructing a particle – not a zebra, not a bicycle, but a particle,” says Kucukelbir – but the current approach doesn’t incorporate that information, and instead works without any “knowledge” of what is being reconstructed. The group plans to explore the possibility of using an existing database of protein structures to build a mathematical model of what particles tend to look like, and incorporate that model into their reconstruction algorithm.
Alp Kucukelbir, Fred Sigworth, Hemant Tagare. A Bayesian Adaptive Basis Algorithm for Single Particle Reconstruction. Journal of Structural Biology. Published online ahead of print.

